News at the platform
"Plenty of fish in the sea" selected image in Biologists@100 conference, JCS - Image Competition
2025-03-27
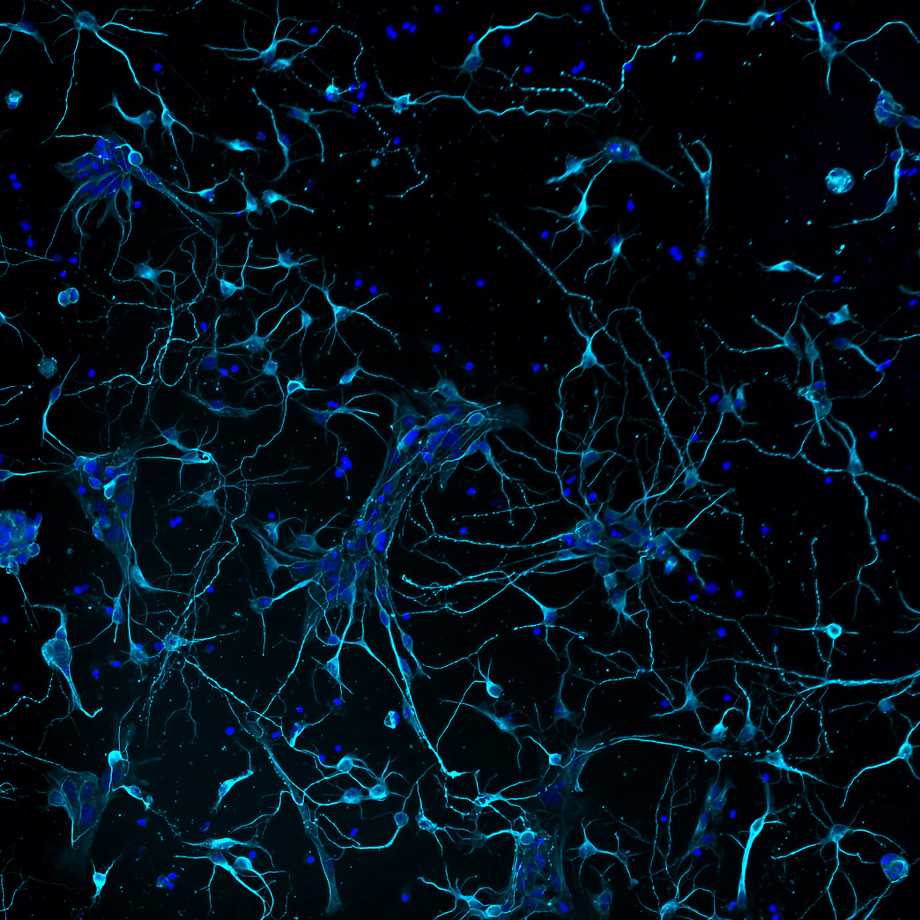
Plenty of fish in the sea - Ludovica Altieri
Murine primary cortical neurons developing interconnections, stained with neuronal tubulin (cyan) and DAPI (blue). This image has been selected among the 15 finalists of the competition organised by The Node and FocalPlane (The Company of Biologists) and was presented at Biologists@100 gallery in Liverpool.
Imaged on a Nikon microscope implemented with a CrestOptics confocal spinning disk module with post-processing using NIS Elements AR by Nikon. Acquired at the IBPM Institute of Molecular Biology and Pathology – CNR National Research Council of Italy, c/o Department of Biology and Biotechnology “Charles Darwin” – Sapienza University of Rome.
FENS Forum 2024" International Neuroscience Conference organized by FENS
2024-06-25
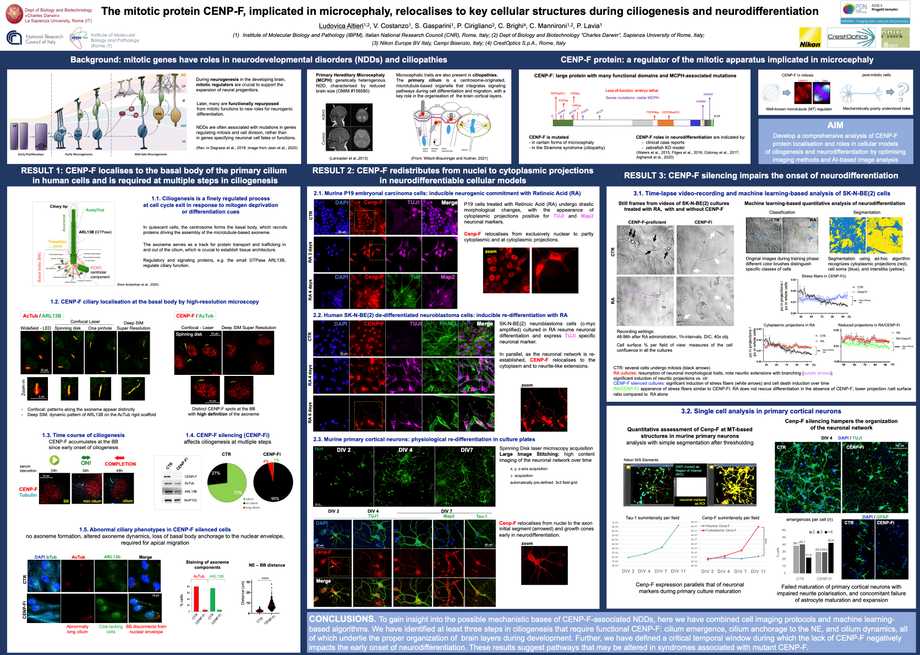
Neurodevelopmental disorders (NDDs) are often associated with mutations in mitotic genes rather than in genes specifying neuronal
functions. Several mitotic regulators are functionally repurposed to new roles during neurodifferentiation. CENP-F, a kinetochore- and microtubule- interacting protein with well-characterised mitotic functions, is being growingly identified for harbouring mutations in primary microcephaly and in the
Strømme syndrome, an NDD classified as a ciliopathy. The mechanism through which mutant CENP-F contributes to these NDDs remains elusive.
To gain insight into the implication of CENP-F in NDDs, we have optimised imaging methods and developed a comprehensive analysis of CENP-F in post- mitotic cellular systems. We first examined CENP-F during the formation of the primary cilium by optimising quantitative immunofluorescence and single- cell analysis of spinning-disk confocal images. CENP-F and some of its interacting partners were characterised in a time-course analysis of ciliogenesis in
human retinal RPE-1 cells. CENP-F localises at the basal body of the cilium and is absolutely required for proper ciliogenesis. We also studied the requirement for CENP-F during neurodifferentiation by time-lapse videorecording and confocal microscopy. We have developed two AI-based algorithms to analyse high-content images of videorecorded cell cultures: that enabled us to extract information on the dynamics with which phenotypic changes occur during neurodifferentiation and identify steps that are sensitive to and/or impaired by CENP-F loss-of-function. In summary, these imaging protocols identify critical temporal windows during which the lack of CENP-F impacts neurodifferentiation and ciliogenesis, thus providing new tools to dissect roles of CENP-F in NDDs.
19th SIBBM Seminar Frontiers in Molecular Biology, The time of molecular biology: development, homeostasis and aging.
2024-06-17
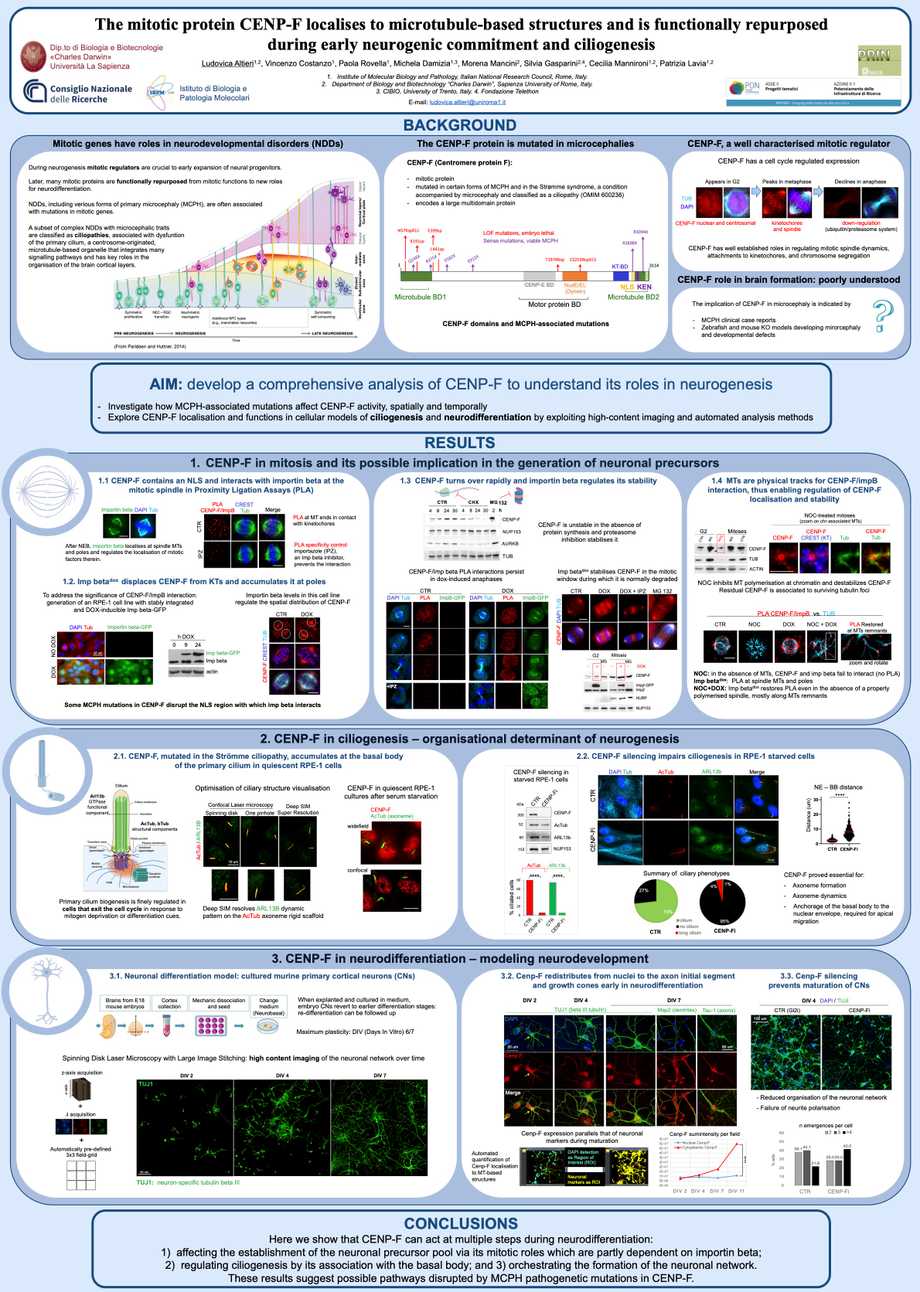
Centromere protein F (CENP-F) is a multidomain protein with an established role in regulating microtubule (MT) functions during mitosis. CENP-F mutations are identified in neurodevelopmental disorders (NDDs) associated with primary microcephaly (MCPH), suggesting that CENP-F may also play roles during commitment to the neurogenic lineage. The molecular mechanism(s) through which mutant CENP-F acts in NDDs however remains elusive. Here we have developed a comprehensive analysis of CENP-F in several cellular systems aiming to understand its roles in neurogenesis. First, we investigated CENP-F C-terminal region, which harbours MCPH mutations. In that region we have characterised a nuclear localisation signal (NLS) flanking a KEN box for ubiquitin-dependent proteasomal degradation. We find that the nuclear import receptor importin beta, which binds the NLS, cooperates with MTs in regulating both CENP-F mitotic localisation and the timing of regulated degradation at mitotic exit. Second, we studied CENP-F in cells that exit the cell cycle and enter quiescence, at which stage they nucleate the primary cilium, a MT-based structure with architectural roles during the organisation of brain cortical layers. We find that CENP-F is required for the axoneme formation and for positioning of the cilium basal body. Third, we characterised CENP-F in neurodifferentiating cellular systems. We identified a CENP-F fraction that mobilises from the nucleus to MT-based structures required for neurite formation and branching. These findings illustrate that CENP-F may act at multiple steps during neurodifferentiation: affecting the establishment of the neuronal precursor pool, the process of ciliogenesis, and the formation of neuronal networks. These results suggest possible pathways disrupted by MCPH pathogenetic mutations in CENP-F.
EMBO Workshop-Visualizing Biological data (VIZBI 2024)
2024-03-15
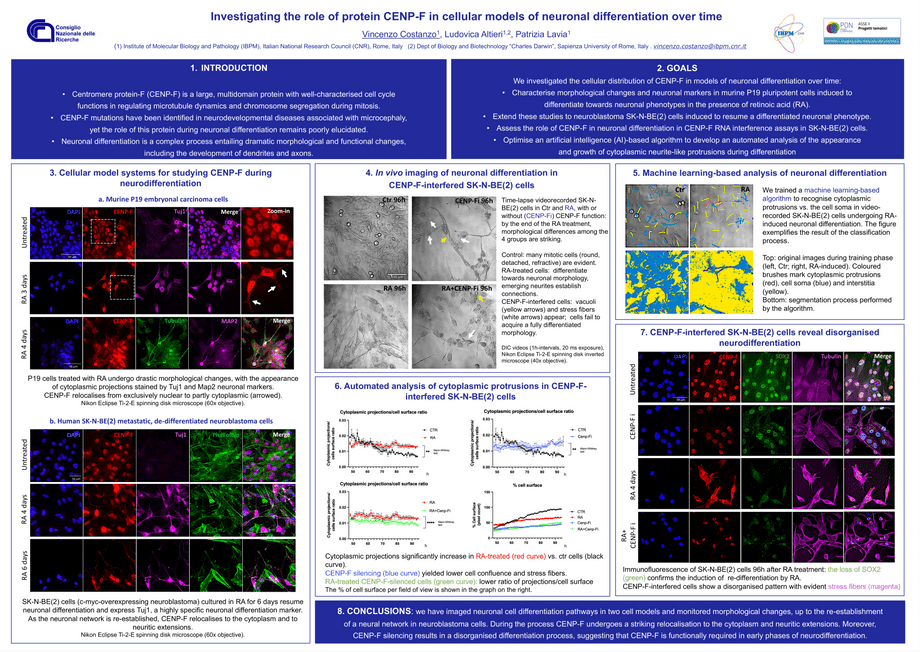
Investigating the role of protein CENP-F in cellular models of neuronal differentiation over time.
Centromere protein-F (CENP-F) is a large multidomain protein with well-characterized functions in regulating microtubule dynamics and chromosome segregation during mitosis. Mutations of CENP-F identified in microcephaly and ciliopathies suggest a critical role of this protein during brain development, which however remains poorly elucidated.
Here, we have investigated CENP-F roles in cellular models of neuronal differentiation. First, we characterised morphological changes and appearance of neuronal markers in murine P19 pluripotent cells induced to differentiate in the presence of retinoic acid. We next extended these studies to dedifferentiated, neuroblastoma SKNBE(2) cells induced to re-establish a differentiated neuronal phenotype. We also performed CENP-F interference assays in SKNBE(2) cells to assess whether the loss of CENP-F would affect neuronal differentiation. The work was assisted by an artificial intelligence-based algorithm that enabled us to perform an automated analysis of differentiation during time-lapse videorecording. We found that CENP-F undergoes a striking relocalisation from the nucleus to the cytoplasm and neuritic extensions during differentiation. CENP-F silencing yields a disorganised differentiation process. These data suggest therefore a requirement for CENP-F in neurodifferentiation.
Imaging 3D cellular models for biomedical applications Theoretical-practical course

2023-10-10
On the 10th-11th of October a microscopy course for the PhD students of the BeMM School will be held at the IBPM-CNR imaging platform (BBCD Department, Polo San Lorenzo).
As detailed in the attached programme, the course will include a theoretical session open to all students; a practical session of confocal spinning disk 3D cell imaging (acquisition and analysis) open to 12 students in presence (divided in subgroups, 2 hours each) and to all interested students from remote. Samples will be provided by the imaging platform but there will be space for questions and discussion where students can explore the suitability of the instrumentation for applications of their interest.
Students who would like to participate in the course are asked to send an email to Giulia Guarguaglini (giulia.guarguaglini@uniroma1.it), specifying whether they would like to attend the practical session in presence. If >12 students will register for an in presence practical session, a selection will be done based on their interests or future plans to use the methods.
EMBO Workshop-VISUALIZING BIOLOGICAL DATA (VIZBI)

2023-03-29
Artificial Intelligence-based analysis of commitment towards neuronal differentiation in living pluripotent cells over time
Neuronal differentiation is a highly complex, multistep process that regulates the determination and specification of neuronal fate, as well as major molecular and functional changes, such as the emergence of neurites and maturation of dendrites and axons. Acquiring a profound knowledge of factors regulating neuronal differentiation is crucial to identify particularly vulnerable windows in which defects or gene mutations can cause neurodevelopmental disorders. This requires capturing the dynamic dimension of the process.
Here we aimed at imaging neuronal differentiation over time, both in fixed cell cultures and under in vivo conditions, to depict morphological cell changes and appearance of neuronal markers. We first characterized differentiating cell models exposed to classical inducers, i.e. Retinoic Acid (RA) and Neurobasal (NB) Medium. We next selected murine P19 pluripotent cells undergoing differentiation for time-lapse recording (up to 96 h) to follow up in real time their commitment to the neuronal fate. We developed two novel artificial intelligence (AI)-based algorithms to perform a high-throughput analysis of the phenotypes elicited during neuronal differentiation.
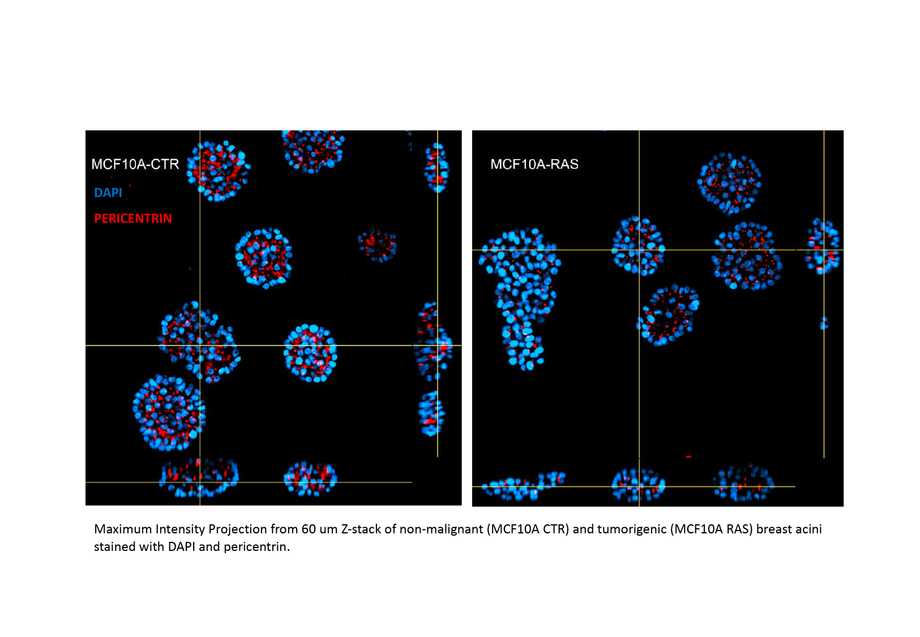
High-content imaging for morphological profiling of 3D breast acini
2022-12-12
Collaborative work between researchers at the IBPM platform and CrestOptics Company allowed the validation of a method for automated imaging and analysis of in vitro 3D breast epithelial acini.
The derived Application Note (https://crestoptics.com/high-content-imaging-for-morphological-profiling-of-3d-breast-acini/) describes high-content platforms, based on the CrestOptics X-Light V3 spinning disk, applied to the morphological profiling of 3D breast spheroids derived from the MCF10A cell line.
The imaging system, coupled with Nis Elements Software capable of automating acquisitions and analyses, has made it possible to compare morphological characteristics between normal (MCF10A CTR) and transformed (MCF10A RAS) acini. High content analysis of morphological features such as volume, elongation, sphericity, diameter, and surface demonstrated that acini derived from transformed cells display an alteration of the acinar structure and architecture (see figure).
The combination of high content microscopy with tridimensional cellular models could thus contribute significantly to cancer research, especially in the validation of active molecules in drug screening.
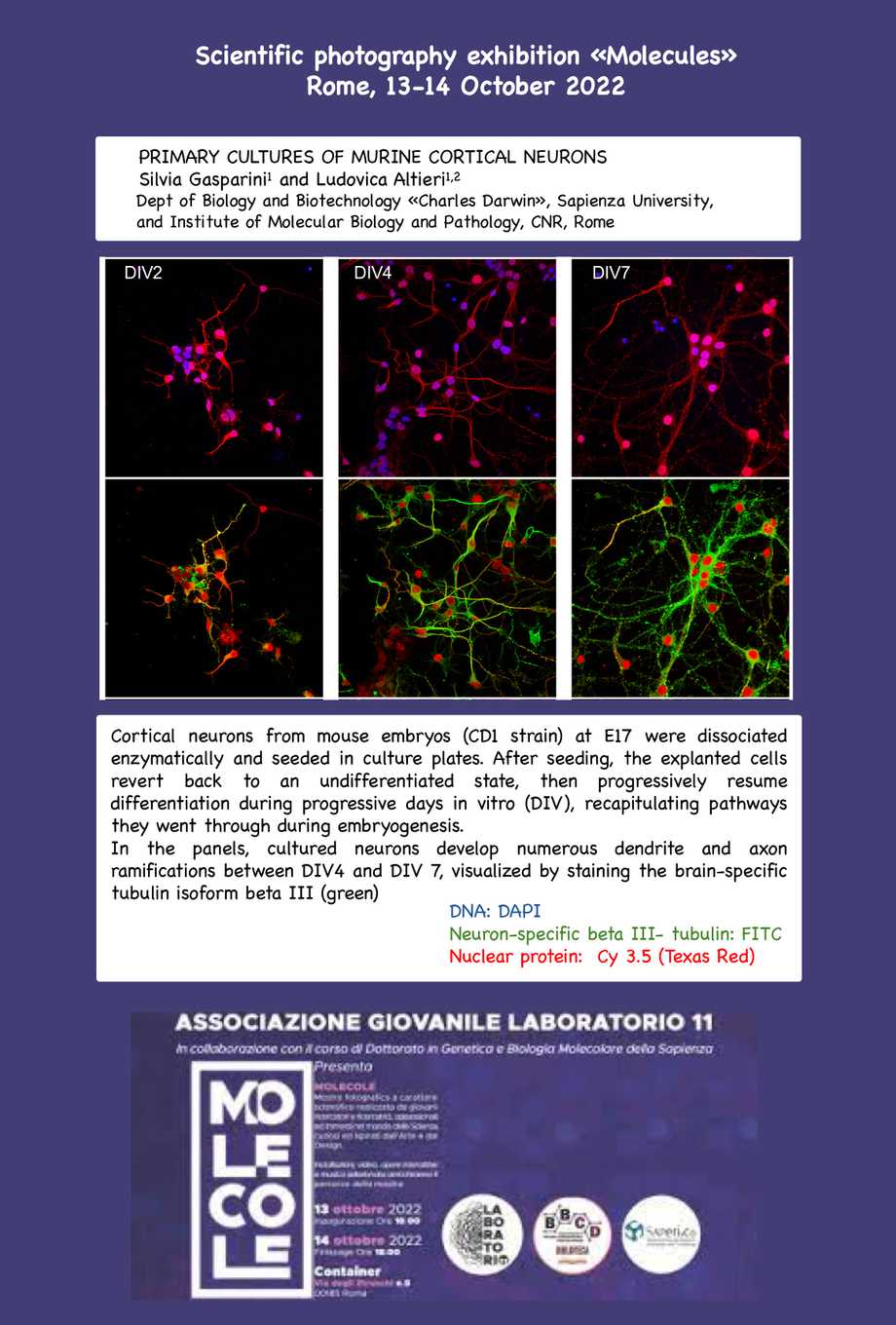
Mostra Molecole
2022-10-13
Images obtained at the Imaging platform were presented at the Exhibition on Scientific Photorgraphy “Molecole”, organized by Associazione Giovanile Laboratorio11 in response to the call “Vitamina G”, in collaboration with the Dept of Biology and Biotechnology Charles Darwin (BBCD), the PhD course in Genetics and Molecular Biologiy of Sapienza University of Rome, the BBCD Department Library, and the inter-departimental Centre “Saperi&Co” of Sapienza.
13-14 October 2022, Container, via degli Etruschi, 15 Roma
Grant award from Regione Lazio to develop 3D high-content imaging methodologies

2021-04-19
The project “Metodologie innovative di 3D high content imaging per la validazione di nuove molecole antitumorali” coordinated by Dr Francesca Degrassi has been funded by Regione Lazio within the Call “Progetti Gruppi di Ricerca 2020”.
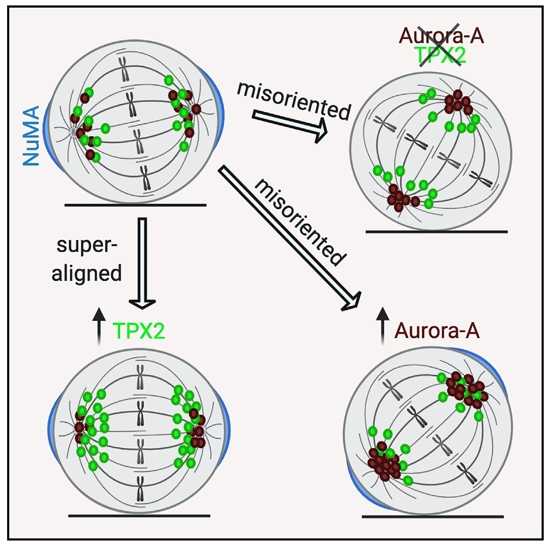
New publication from the microscopy platform
2021-02-08
Polverino F, Naso FD, Asteriti IA, Palmerini V, Singh D, Valente D, Bird AW, Rosa A, Mapelli M, Guarguaglini G. The Aurora-A/TPX2 Axis Directs Spindle Orientation in Adherent Human Cells by Regulating NuMA and Microtubule Stability.
Curr Biol. 2021. 31:658-667.e5.
doi: 10.1016/j.cub.2020.10.096. Epub 2020 Dec 3.
In this study we characterised the role of the Aurora-A kinase and its major regulator TPX2 in the control of mitotic spindle orientation in mammalian cells, a process which determines the position and fate of daughter cells.
Work was collaboratively carried out by groups at the Institute of Molecular Biology and Pathology (IBPM) and at the European Institute of Oncology (IEO). Research groups at Sapienza University and at the Max Planck Institute of Molecular Physiology (Dortmund, DE) participated to the project.
IBPM reference person: Giulia Guarguaglini
Events
Visualizing biological interactions in cells and cellular compartments

2024-10-21
The 3rd edition of the theoretical and practical imaging course for the BEEM PhD students will be offered at the Imaging Platform of the CNR Institute of Molecular Biology and Pathology.
Visualizing biological interactions in cells and cellular compartments

2024-10-21
The 3rd edition of the theoretical and practical imaging course for the BEEM PhD students will be offered at the Imaging Platform of the CNR Institute of Molecular Biology and Pathology.
3D high content imaging workshop

2024-06-05
On the 5th of July the final workshop of the Regione Lazio project "3D high content imaging for the validation of new anti-cancer molecules" will be held at the BBCD Department, Polo San Lorenzo (Aula Franco Tatò, Via dei Sardi 70, II floor). As detailed in the attached programme, the workshop gathers together speakers from research institutions, universities and companies with the aim of providing an overview of innovative methodologies in 3D cellular imaging for biomedical applications and drug screening.
To participate please send an email to Giulia Guarguaglini (giulia.guarguaglini@uniroma1.it) or Francesca Degrassi (francesca.degrassi@uniroma1.it). Attendance will be limited to 60 participants.
Cellular microscopy and image analysis - Theoretical-practical course

2022-07-13
A theoretical-practical course was offered at the Imaging platform for PhD students from 9 PhD courses associated to the PhD School in Molecular Biology and Medicine at Sapienza University of Rome.
Seminars in: Research Design in Genetics and Molecular Biology, Sapienza University

2021-04-09
An account of how coupling high-resolution imaging metods with mathematics and physical modelling provides insight into cell dynamics.
Guest speaker: Emanuele Roscioli, Fondazione Toscana Life Sciences, Siena.
Incontri al DSB – EuroBioImaging Italia and its nodes

2021-04-06
Euro-BioImaging is the European landmark research infrastructure for biological and biomedical imaging. Through Euro-BioImaging, life scientists can access imaging instruments, expertise, training opportunities and data management services. The webinar presented the activity of the Italian nodes to CNR biomedical scientists.
Workshop – Artificial intelligence and automation meet cellular imaging in biomedical research
2021-03-25
Cellular imaging is undergoing an unprecedented development that has greatly amplified its informative power with the introduction of machine learning, artificial intelligence-based approaches and quantitative data analysis. The workshop has explored the application of these methods in as diverse cell biology fields as cell migration a, morphological reconstruction of subcellular structures, and use of model organism- and cell-based assays in genetic and drug screening.