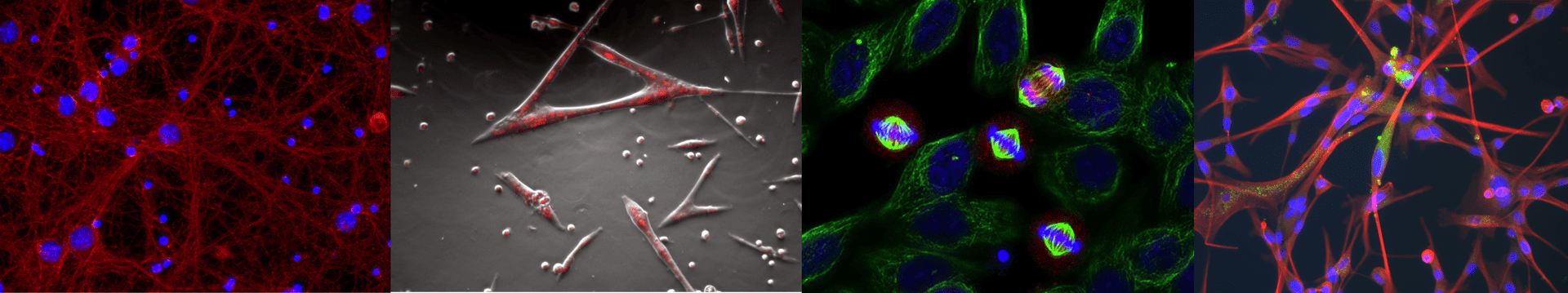
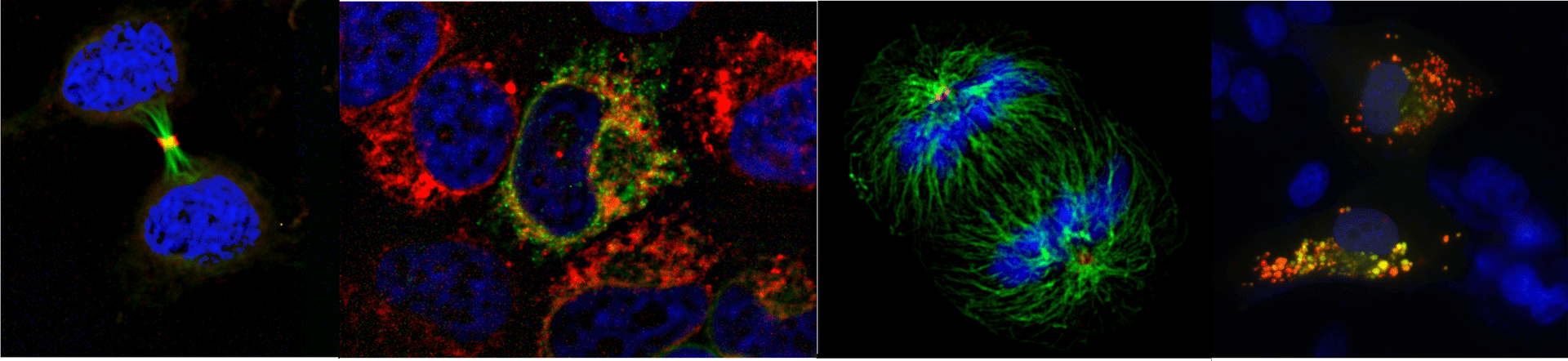
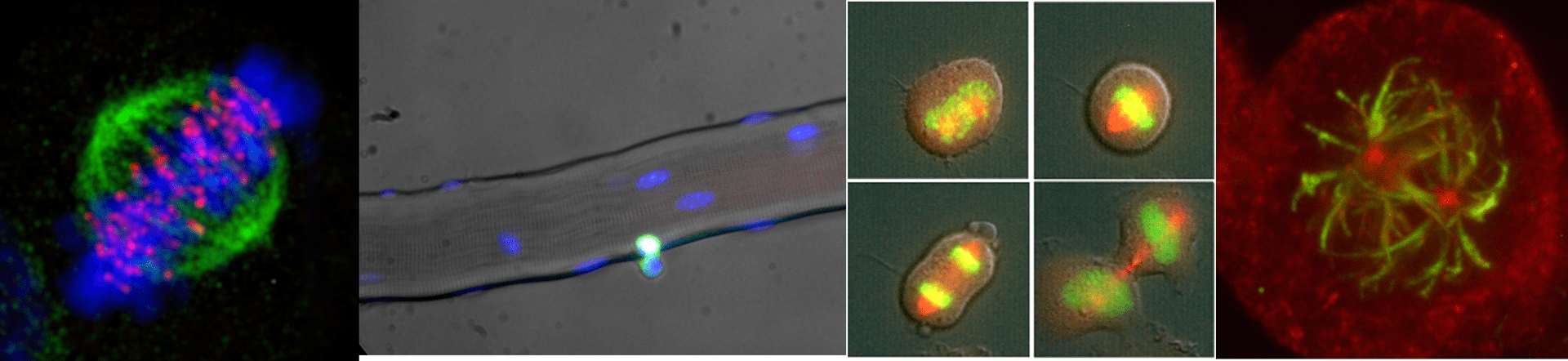
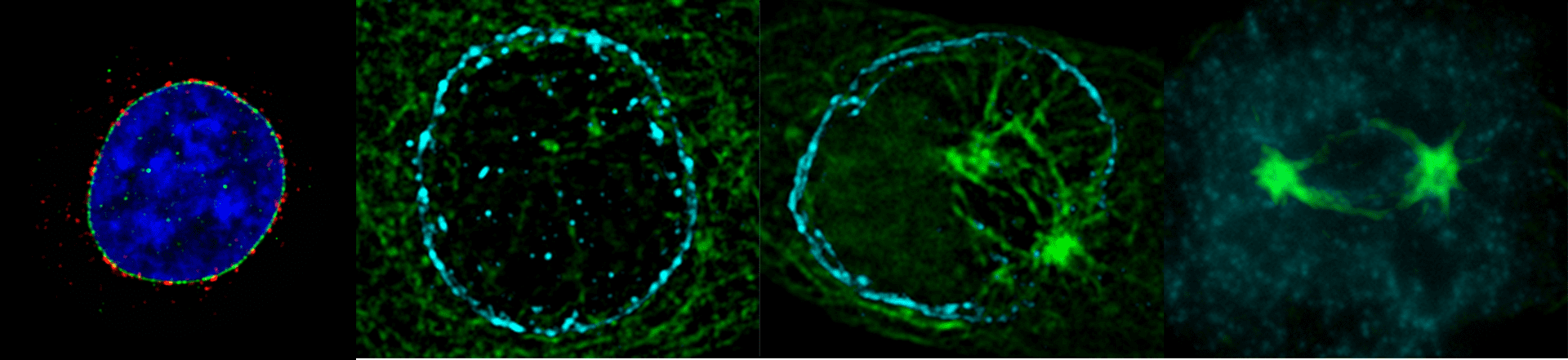
Imaging platform
Institute of Molecular Biology and Pathology

Visualizing cell dynamics
The IBPM microscopy platform hosts last-generation widefield light microscopy equipment and advanced imaging software for automated image analyses.
It supports versatile applications for both fixed and live cell samples and provides opportunities for coupling dynamic studies (time-lapse recording) with high resolution qualitative and quantitative image analysis.
The Center is open to external users, click here for access rules.
Applications
- Time-lapse recording of cellular processes in live cells over days
- Detecting intracellular molecular interactions
- Reconstructing cellular and subcellular structures at 3D level
- Quantitative analysis of imaging data
About us
The platform was first established in 2009 with support from Fondazione Monte Paschi, Fondazione Roma and Assicurazioni Generali S.p.A. It has since been supported by two dedicated grants from Regione Lazio and by the CNR Project “Impara - Imaging from Molecules to Preclinical Studies” funded by the Ministry of Education and Research for the development of National research infrastructures. The platform is hosted at the Department of Biology and Biotechnology “Charles Darwin” of Sapienza University.
Latest News
"Plenty of fish in the sea" selected image in Biologists@100 conference, JCS - Image Competition
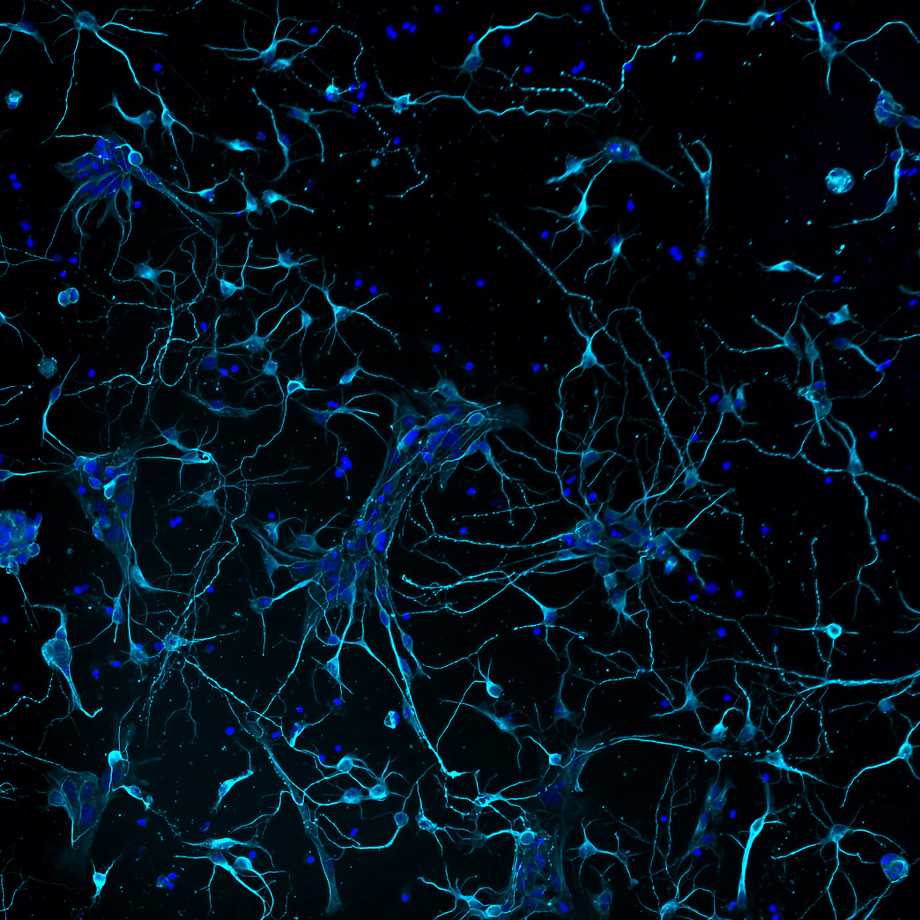
Plenty of fish in the sea - Ludovica Altieri
Murine primary cortical neurons developing interconnections, stained with neuronal tubulin (cyan) and DAPI (blue). This image has been selected among the 15 finalists of the competition organised by The Node and FocalPlane (The Company of Biologists) and was presented at Biologists@100 gallery in Liverpool.
Imaged on a Nikon microscope implemented with a CrestOptics confocal spinning disk module with post-processing using NIS Elements AR by Nikon. Acquired at the IBPM Institute of Molecular Biology and Pathology – CNR National Research Council of Italy, c/o Department of Biology and Biotechnology “Charles Darwin” – Sapienza University of Rome.
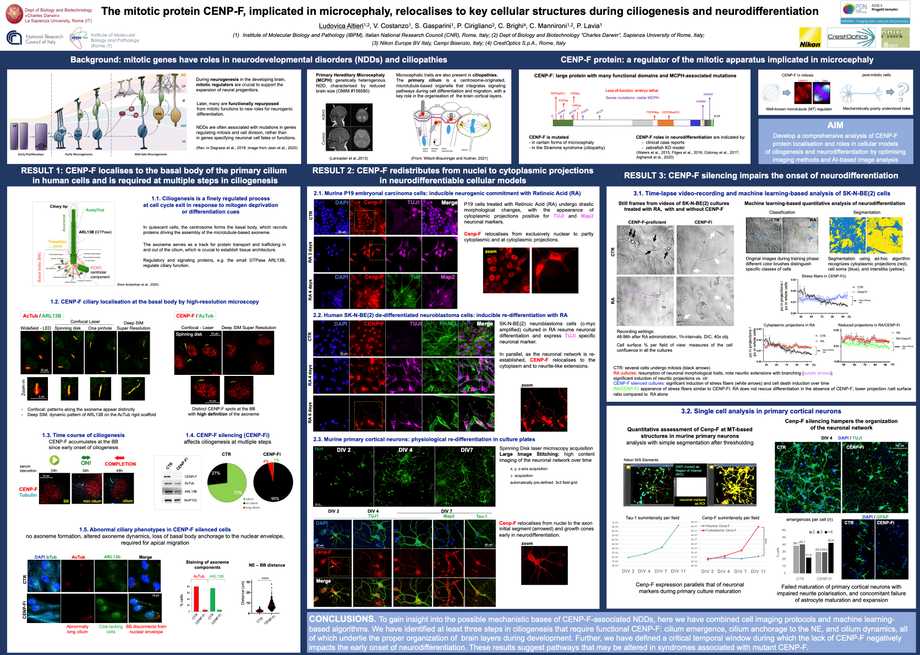
FENS Forum 2024" International Neuroscience Conference organized by FENS
Neurodevelopmental disorders (NDDs) are often associated with mutations in mitotic genes rather than in genes specifying neuronal
functions. Several mitotic regulators are functionally repurposed to new roles during neurodifferentiation. CENP-F, a kinetochore- and microtubule- interacting protein with well-characterised mitotic functions, is being growingly identified for harbouring mutations in primary microcephaly and in the
Strømme syndrome, an NDD classified as a ciliopathy. The mechanism through which mutant CENP-F contributes to these NDDs remains elusive.
To gain insight into the implication of CENP-F in NDDs, we have optimised imaging methods and developed a comprehensive analysis of CENP-F in post- mitotic cellular systems. We first examined CENP-F during the formation of the primary cilium by optimising quantitative immunofluorescence and single- cell analysis of spinning-disk confocal images. CENP-F and some of its interacting partners were characterised in a time-course analysis of ciliogenesis in
human retinal RPE-1 cells. CENP-F localises at the basal body of the cilium and is absolutely required for proper ciliogenesis. We also studied the requirement for CENP-F during neurodifferentiation by time-lapse videorecording and confocal microscopy. We have developed two AI-based algorithms to analyse high-content images of videorecorded cell cultures: that enabled us to extract information on the dynamics with which phenotypic changes occur during neurodifferentiation and identify steps that are sensitive to and/or impaired by CENP-F loss-of-function. In summary, these imaging protocols identify critical temporal windows during which the lack of CENP-F impacts neurodifferentiation and ciliogenesis, thus providing new tools to dissect roles of CENP-F in NDDs.
Members
 Lia Asteriti
Lia Asteritilia.asteriti@uniroma1.it
 Francesca Degrassi
Francesca Degrassifrancesca.degrassi@uniroma1.it
 Giulia Fianco
Giulia Fiancogiulia.fianco@gmail.com
 Giulia Guarguaglini
Giulia Guarguaglinigiulia.guarguaglini@uniroma1.it
 Patrizia Lavia
Patrizia Laviapatrizia.lavia@uniroma1.it
 Valerio Licursi
Valerio Licursivalerio.licursi@uniroma1.it
 Federica Polverino
Federica Polverinofedericapolverino@cnr.it
 Davide Valente
Davide Valentedavide.valente@cnr.it
 Ludovica Altieri
Ludovica Altieriludovicaaltieri@cnr.it
Sponsors